Preprocessing dietary and CGM data
Heather Hall
2015-08-05
Last updated: 2017-09-13
Code version: 2c087f8
Load libraries
library(lubridate) # work with times
library(leaps) # regsubset
library(kknn) # k nearest neighbor
library(gbm) # boosting
library(randomForest) # random forest
library(gridExtra); library(grid); library(gtable) # plot tables
library(ggplot2) # ggplot
library(scales) # easy plotting of dates
# load personal library
source(paste(personal.library.directory, "project_library.R", sep = "/"))Read in files
# read in myfitnesspal data and recorded meal timing
food.logs = readMyFitnessPal(food.data.path, conversion.chart.path)
food.times <- read.delim(food.timing.file, stringsAsFactors = FALSE)
# read in glucose data
cgm.readings = readDexcomFile(glucose.file)preview of CGM data
head(cgm.readings)
GlucoseDisplayDate GlucoseDisplayTime GlucoseValue DisplayTime
1 3/29/15 11:53 100 2015-03-29 11:53:00
2 3/29/15 11:58 111 2015-03-29 11:58:00
3 3/29/15 12:03 113 2015-03-29 12:03:00
4 3/29/15 12:08 114 2015-03-29 12:08:00
5 3/29/15 12:13 116 2015-03-29 12:13:00
6 3/29/15 12:18 121 2015-03-29 12:18:00
example.date = "3/30/15"
example.CGM = cgm.readings[cgm.readings$GlucoseDisplayDate == example.date, ]
example.CGM$GlucoseDisplayTime <- as.POSIXct(strptime(example.CGM$GlucoseDisplayTime, format="%H:%M"))
ggplot(example.CGM) +
aes(x = GlucoseDisplayTime, y = GlucoseValue, group = GlucoseDisplayDate) +
geom_point() + geom_line() +
scale_x_datetime(breaks = date_breaks("6 hour"), labels = date_format("%H:%M")) +
labs(x = "time of day (HH:MM)", y = "interstitial glucose (mg/dL)")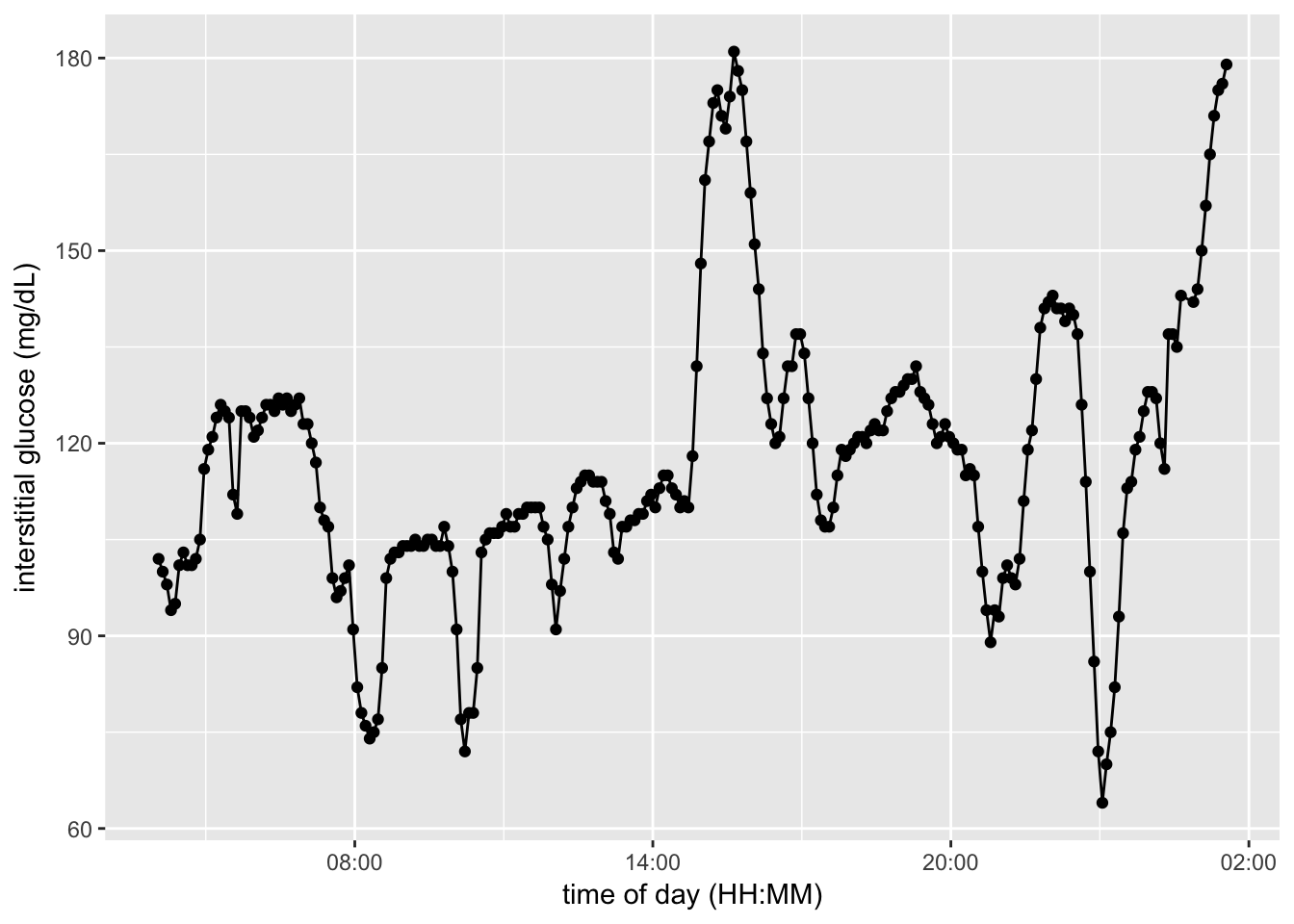
preview of food logs
current.table = food.logs[sample(1:nrow(food.logs), 10), ]
head(current.table)
description food_id entry_date category
2712 White Rice 169888484 1/29/15 <NA>
2356 Rice - White medium-grain cooked 219188109 8/3/15 Dinner
2742 Whole Psyllium Husk 212702230 5/21/16 Dinner
1412 Garlic & Herb Cheese 131419740 12/19/14 <NA>
515 Cereal 160355225 3/29/15 Breakfast
1005 Coffee - Brewed from grounds 155741288 1/9/15 <NA>
amount calories total_fat
2712 1 cup 205.00 0.40000
2356 1 cup 169.26 0.27342
2742 2.5 serving(s) of 0.5 tablespoon (4g) 37.50 NA
1412 1 wedge 50.00 3.00000
515 1 cup 200.00 4.00000
1005 1 cup (8 fl oz) 4.74 0.09480
polyunsaturated_fat monounsaturated_fat trans_fat cholesterol sodium
2712 0.100000 0.10000 0 0 2.00
2356 0.072912 0.08463 0 0 0.00
2742 NA NA NA NA NA
1412 NA NA NA 15 220.00
515 1.000000 1.00000 0 0 280.00
1005 0.004740 0.07110 0 0 9.48
potassium carbs fiber sugar protein vitamin_a vitamin_c calcium
2712 55.000 45.00000 0.6000 0.1 4.20000 0 0 0.0000
2356 37.758 37.22418 0.3906 0.0 3.09876 0 0 0.3906
2742 NA 10.00000 10.0000 NA NA NA NA NA
1412 NA 0.50000 0.0000 0.0 3.00000 2 0 11.0000
515 360.000 40.00000 6.0000 2.0 6.00000 20 20 20.0000
1005 232.260 0.00000 0.0000 0.0 0.56880 0 0 0.9480
iron Food.group food species
2712 2.0000000 grain rice
2356 10.7776694 grain rice
2742 NA fiber psyllium plantago
1412 0.0000000 dairy,vegetable cheese,garlic
515 90.0000000 grain,grain,grain,grain wheat,corn,oat,rice
1005 0.2633335 drink coffee
additional_desc
2712
2356
2742
1412
515 Cereal life by quaker
1005 preview of food timing and meal descriptions
head(food.times[sample(1:nrow(food.times), 10), ])
Date TIME AM_PM Food Meal Minus.Meal Notes
31 4/3/15 17:20 pm wine dinner NA
149 4/27/15 19:30 pm dinner dinner NA unsure
1 3/29/15 9:15 am Banana breakfast NA
79 4/13/15 7:31 am breakfast breakfast NA
93 4/15/15 12:05 pm lunch lunch NA
77 4/12/15 14:55 pm mandarin orange NA
description
31 Wine-cabernet Savignon
149
1 Bananas - Raw
79
93
77 ClematineAnnotate glucose spikes
min.peak = 120
min.peak.height = 30
cgm.readings = markGlucosePeaks(cgm.readings, min.peak.height, min.peak)
# count number of peaks
peak.type = c("max")
number.peaks = length(cgm.readings[which(cgm.readings$peak %in% peak.type),]$peak)
print(paste(number.peaks, "total glucose spikes annotated"))[1] "261 total glucose spikes annotated"preview of annotated glucose spikes
head(cgm.readings)
GlucoseDisplayDate GlucoseDisplayTime GlucoseValue DisplayTime
1 3/29/15 11:53 100 2015-03-29 11:53:00
2 3/29/15 11:58 111 2015-03-29 11:58:00
3 3/29/15 12:03 113 2015-03-29 12:03:00
4 3/29/15 12:08 114 2015-03-29 12:08:00
5 3/29/15 12:13 116 2015-03-29 12:13:00
6 3/29/15 12:18 121 2015-03-29 12:18:00
peak timeChange glucoseChange
1 0 0 0
2 min 5 111
3 0 0 0
4 0 0 0
5 0 0 0
6 0 0 0
# get same example date
example.CGM = cgm.readings[cgm.readings$GlucoseDisplayDate == example.date, ]
example.CGM$GlucoseDisplayTime <- as.POSIXct(strptime(example.CGM$GlucoseDisplayTime, format="%H:%M"))
# show example of labeled cgm data
spike.colors = c("0" = "black", "max" = "red", "min" = "purple")
ggplot(example.CGM) +
aes(x = GlucoseDisplayTime, y = GlucoseValue, group = GlucoseDisplayDate) +
geom_point(aes(color = peak)) + geom_line() +
scale_x_datetime(breaks = date_breaks("6 hour"), labels = date_format("%H:%M")) +
scale_colour_manual(values = spike.colors) +
labs(x = "time of day (HH:MM)", y = "interstitial glucose (mg/dL)")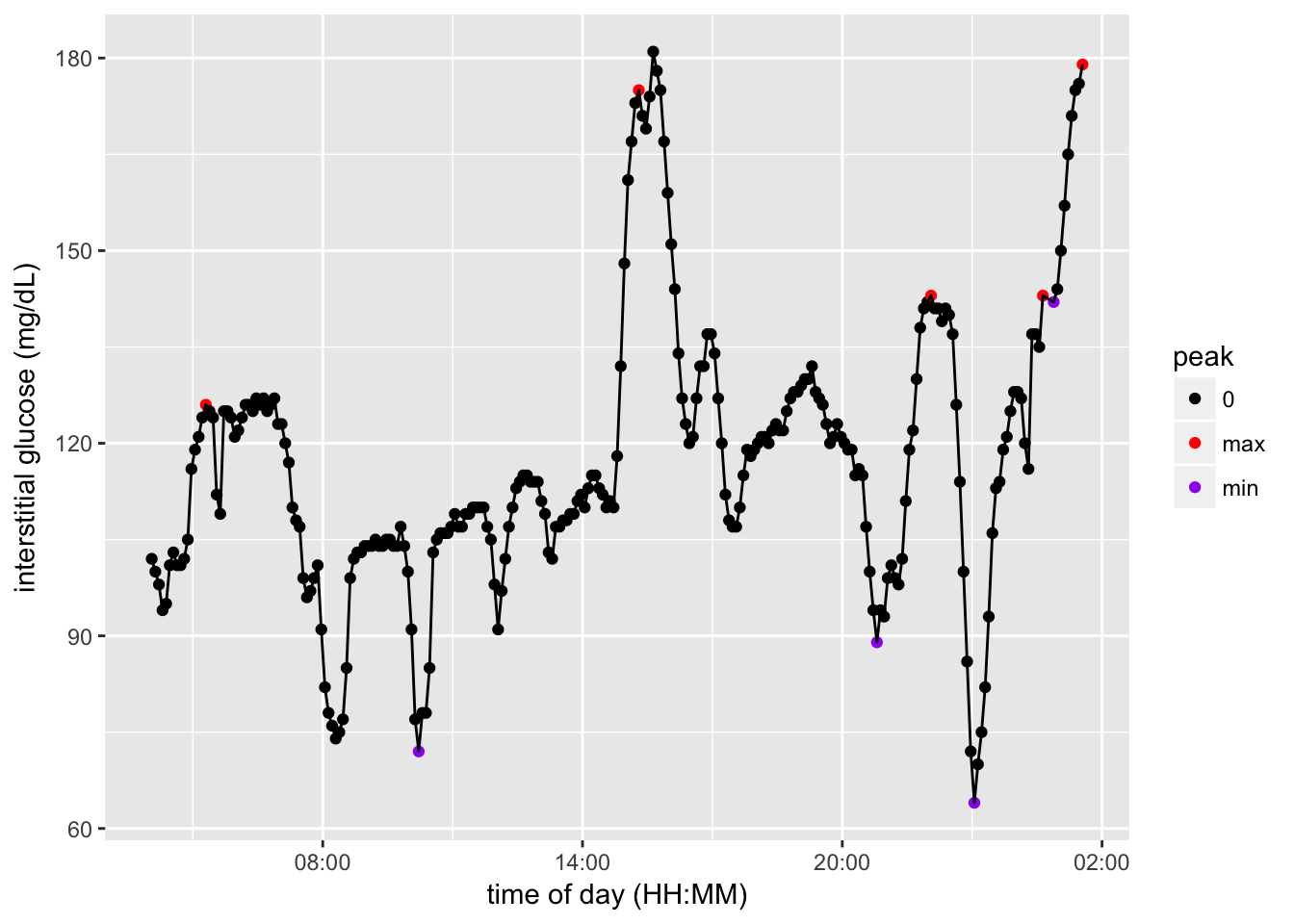
distribution of glucose concentration
peakLabeller <- as_labeller(c(
"0" = "all data",
"max" = "spikes (max)",
"min" = "nadirs (min)"
))
ggplot(cgm.readings) +
aes(x = GlucoseValue, group = peak, fill = peak) +
geom_density(alpha = 0.3) +
ggtitle("Distribution of interstitial glucose concentrations") +
facet_wrap(~peak, labeller=peakLabeller) +
theme(legend.position="none")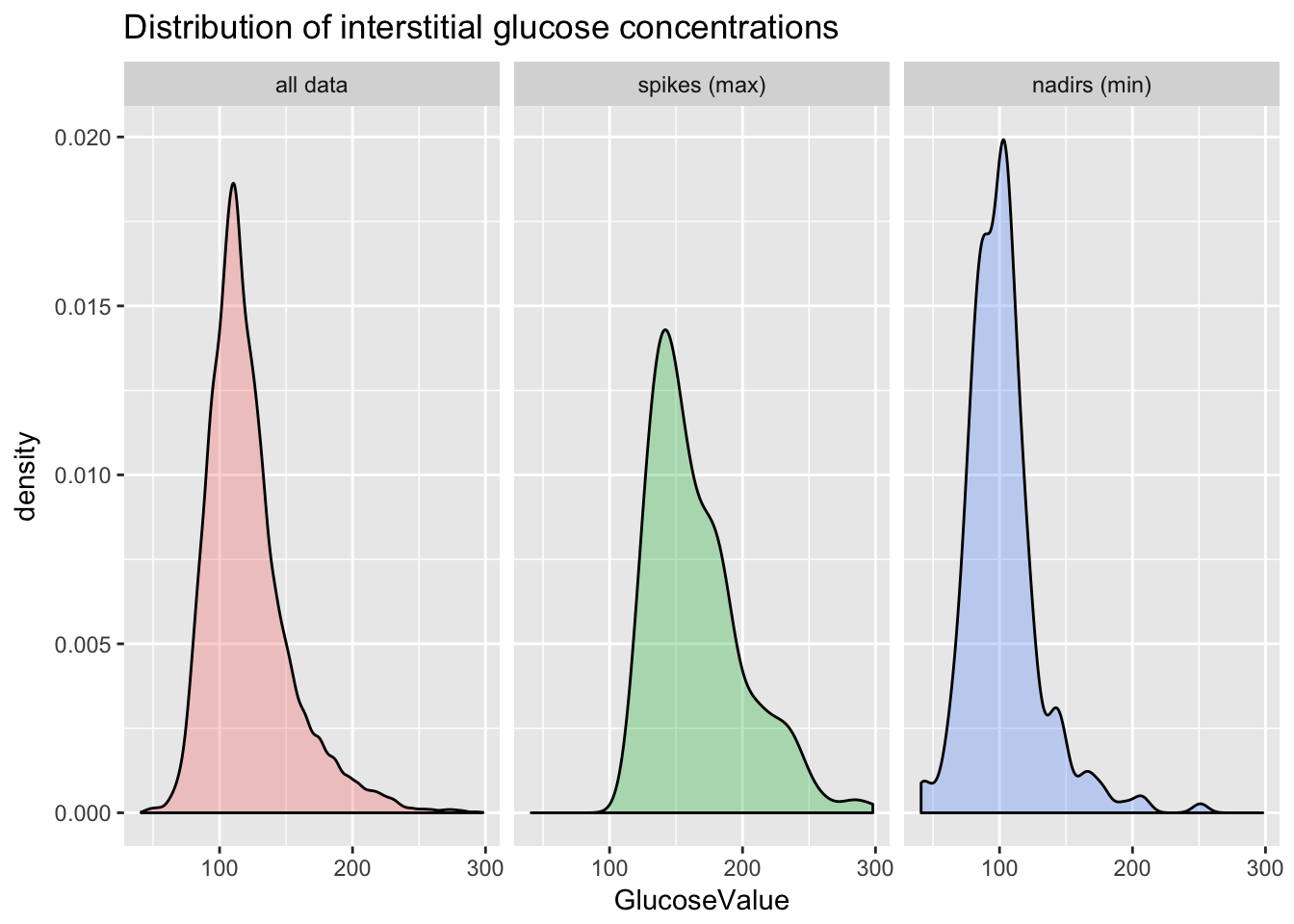
Data exclusion
# keep only glucose spikes
# --------------------------
cgm.readings = cgm.readings[cgm.readings$peak == peak.type,]
# keep only dates with recorded meal times
# --------------------------------
dates.with.meal.times <- unique(food.times$Date)
cgm.readings <- cgm.readings[which(cgm.readings$GlucoseDisplayDate %in% dates.with.meal.times),]
# count number of peaks
peaks.remaining = length(cgm.readings[which(cgm.readings$peak %in% peak.type),]$peak)
print(paste(peaks.remaining, "peaks remaining with meal data"))[1] "201 peaks remaining with meal data"# remove travel from glucose data and food timing
# --------------------------------
cgm.readings = removeTravelDates(cgm.readings, travel.file, TZ.path)
food.times = food.times[food.times$Date %in% cgm.readings$GlucoseDisplayDate, ]
# print number of days removed from travel
peaks.post.travel = length(cgm.readings[which(cgm.readings$peak %in% peak.type),]$peak)
print(paste(peaks.post.travel, "peaks remaining after excluding travel"))[1] "157 peaks remaining after excluding travel"Session information
sessionInfo()R version 3.3.3 (2017-03-06)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X El Capitan 10.11.6
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid parallel splines stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] scales_0.4.1 ggplot2_2.2.1 gtable_0.2.0
[4] gridExtra_2.2.1 randomForest_4.6-12 gbm_2.1.3
[7] lattice_0.20-35 survival_2.41-3 kknn_1.3.1
[10] leaps_3.0 lubridate_1.6.0
loaded via a namespace (and not attached):
[1] igraph_1.0.1 Rcpp_0.12.10 knitr_1.17 magrittr_1.5
[5] munsell_0.4.3 colorspace_1.3-2 plyr_1.8.4 stringr_1.2.0
[9] tools_3.3.3 git2r_0.19.0 htmltools_0.3.5 lazyeval_0.2.0
[13] yaml_2.1.14 rprojroot_1.2 digest_0.6.12 tibble_1.3.0
[17] Matrix_1.2-8 evaluate_0.10.1 rmarkdown_1.6 labeling_0.3
[21] stringi_1.1.5 backports_1.0.5 This R Markdown site was created with workflowr